Generating Jupyter Notebooks#
After setting up a workflow using the Napari Assistant, we can export Python code, e.g. as Jupyter Notebook.
This tutorial is also available as video export_notebooks.mp4
In the Assistant panel, click on the Generate Code... button and the Export Jupyter Notebook using Napari menu.
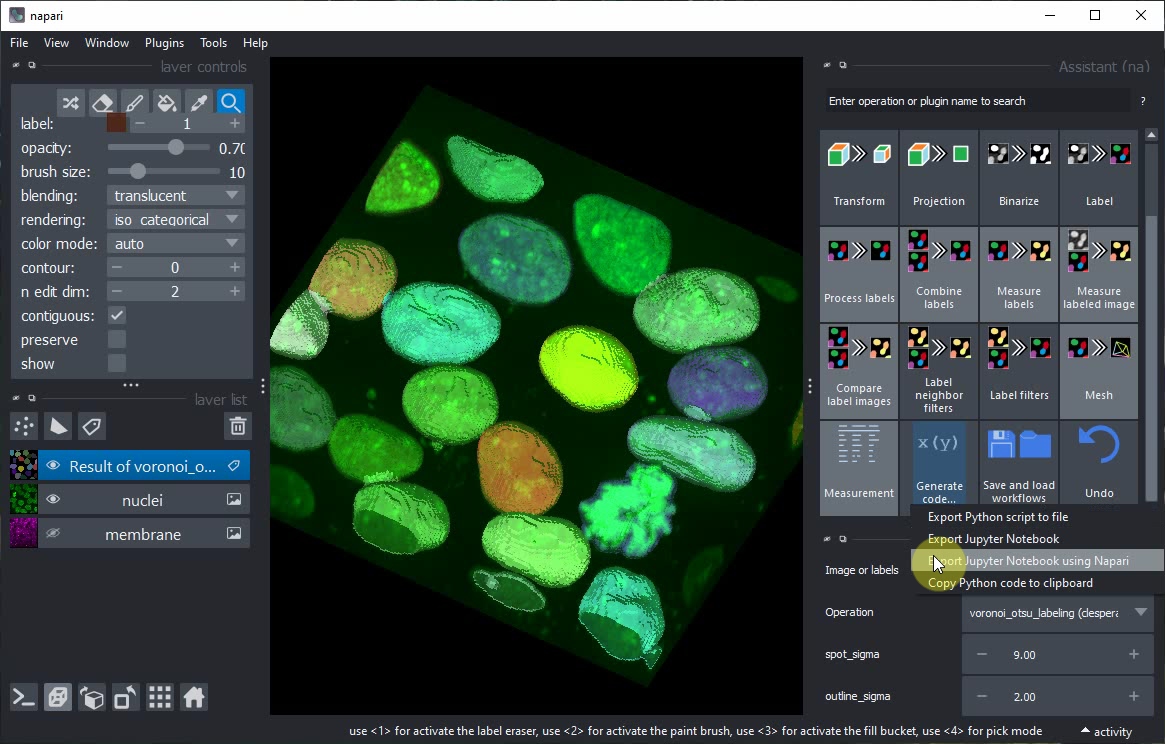
Jupyter lab will open and ask you to select a Kernel. Keep the default option and click on Select.

When executing the notebook, errors may appear, e.g. when loading the data.

Scroll down to the end of the error message to read what didn’t work.

Scoll up again to the notebook cell that didn’t work and modify the code so that it uses the imread function to load the image from disc.
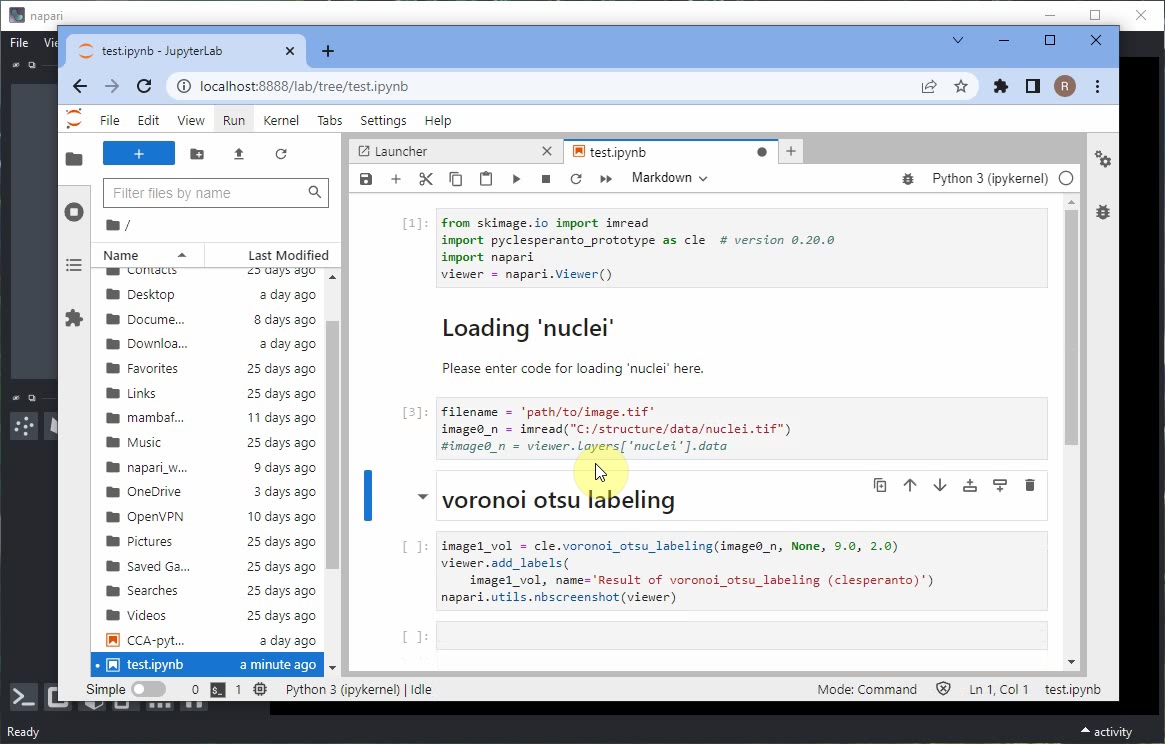
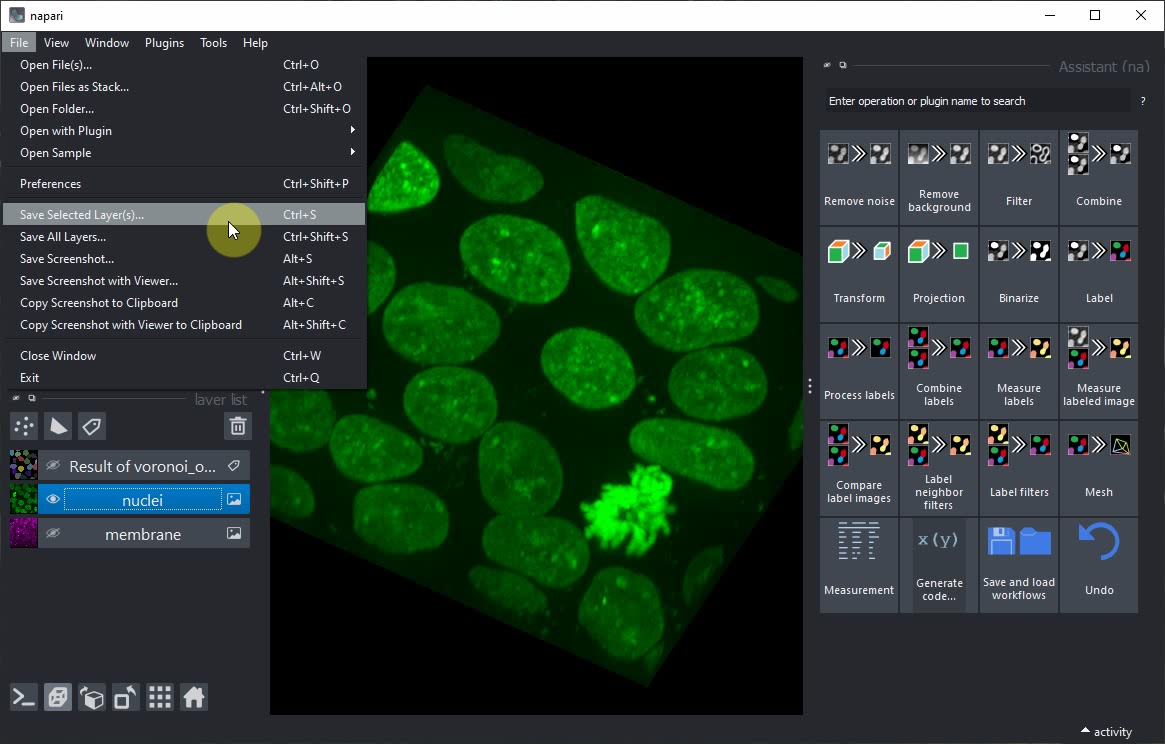
In case you don’t have the nuclei image saved on disc already, use the menu File > Save current Layer(s) to save the nuclei layer as .tif file.
Afterwards, rerun the notebook and inspect the result. The Napari viewer that opened in the background will also be shown within the notebook.
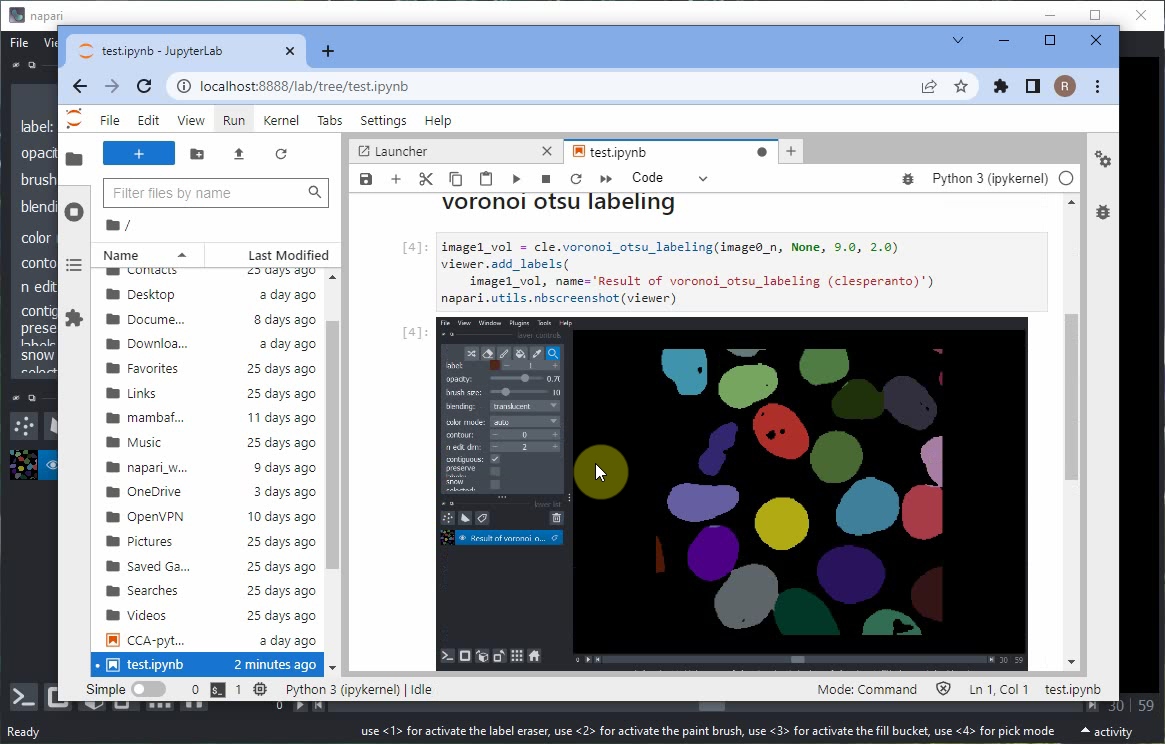
If you also want to view the raw image data together with the segmentation result, add these lines to your code:
viewer.add_image(image0_n)
napari.utils.nbscreenshot(viewer)

Rerun the notebook, or modify the order of layers manually in the Napari viewer. At the end it should look like this.
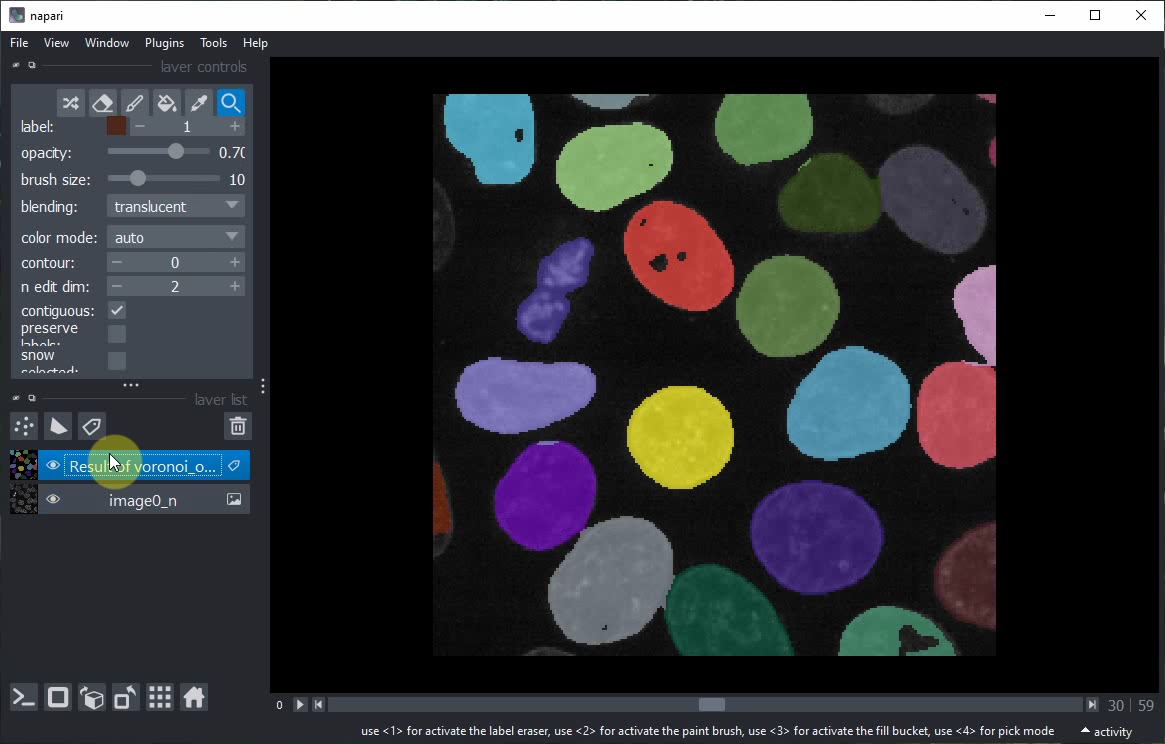
Voila! You have now generated a Jupyter Notebook from a Napari Assistant workflow. This notebook documents your work reproducibly and can be shared with others.
Exercise#
Load the image human_mitosis from the menu File > Open Samples > napari builtins > Human mitosis and design a workflow for segmenting the nuclei. Export the workflow as Jupyter notebook.
