Data Transformation#
This notebook focuses on transforming cleaned data into analysis-ready features.
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler, StandardScaler
patients_cleaned = pd.read_csv("data/patients_cleaned.csv")
patients_cleaned.head()
| patient_id | age | admission_date | discharge_date | gender | weight | height | blood_pressure | admission_unit | albumin_g_dl | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 39.0 | 2023-01-05 | 2023-01-10 | f | 78.0 | 184.0 | 130.0 | Surgery | 1.00 |
| 1 | 2 | 60.0 | 2023-02-11 | 2023-02-20 | m | 55.0 | 180.0 | 90.0 | Cardiol. | 4.26 |
| 2 | 4 | 50.0 | 2023-03-20 | 2023-03-25 | f | 70.0 | 179.0 | 110.0 | Intensive Care U. | 3.10 |
| 3 | 10 | 45.0 | 2023-07-05 | 2023-07-15 | m | 90.0 | 165.0 | 80.0 | Neurology | 3.10 |
| 4 | 12 | 90.0 | 2023-08-10 | 2023-08-20 | female | 45.0 | 170.0 | 60.0 | Intensive Care U. | 4.26 |
#patients_cleaned.info()
1 Checking categorical variables#
Categorical variables often contain multiple spellings or encodings for the same concept. We need to standardize these.
Here we focus on the
gendercolumn:First, we inspect unique values and their counts.
Then we replace inconsistent entries with a standard value.
Casting to
categorydtype can reduce memory and make intent explicit.
# Show number of unique categories and their counts
patients_cleaned['gender'].value_counts(dropna=False)
gender
m 9
male 9
f 6
female 2
Name: count, dtype: int64
# Correct inconsistent entries - f -> female, m -> male
patients_cleaned['gender'] = patients_cleaned['gender'].replace('f', 'female')
patients_cleaned['gender'] = patients_cleaned['gender'].replace('m', 'male')
patients_cleaned['gender'].value_counts(dropna=False)
gender
male 18
female 8
Name: count, dtype: int64
# visualize gender distribution
Check admission_unit for unique values#
# Check unique values in admission_unit
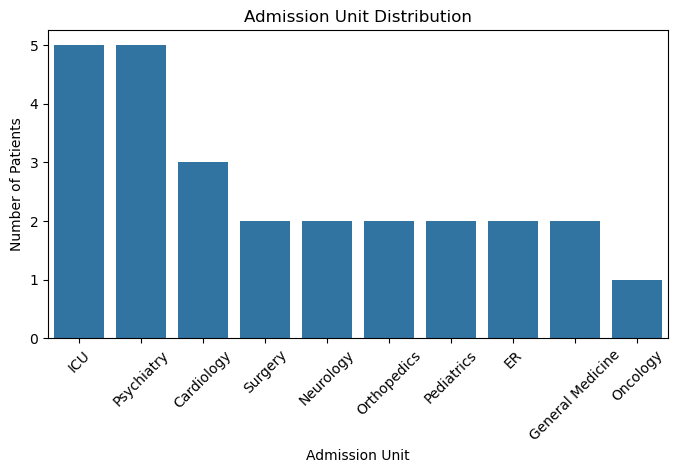
patients_cleaned['admission_unit'].value_counts(dropna=False)
admission_unit
Intensive Care U. 5
Psychiatry 5
Cardiol. 3
Surgery 2
Neurology 2
Orthopedics 2
Pediatrics 2
Emergency Room 2
General Medicine 2
Oncology 1
Name: count, dtype: int64
Rename inconsistent entries#
# Standardize admission_unit entries
patients_cleaned['admission_unit'] = patients_cleaned['admission_unit'].replace({'Emergency Room': 'ER', 'Intensive Care U.': 'ICU', 'Cardiol.': 'Cardiology'})
# visualize admission_unit distribution
plt.figure(figsize=(8,4))
sns.countplot(data=patients_cleaned, x='admission_unit', order=patients_cleaned['admission_unit'].value_counts().index)
plt.title('Admission Unit Distribution')
plt.ylabel('Number of Patients')
plt.xlabel('Admission Unit')
plt.xticks(rotation=45)
plt.show()
2 Encoding categorical variables for modeling#
Many models require numeric inputs.
pd.get_dummies()creates one-hot encoded columns for categorical variables.For high-cardinality categorical features you may want alternative encoding strategies (target encoding, embedding, hashing).
One-hot encoding for ‘gender’#
# One-hot encode gender
patients_cleaned = pd.get_dummies(patients_cleaned, columns=['gender'], drop_first=True)
Label Encoding for ‘admission_unit’#
For admission_unit with multiple categories, we use label encoding to convert categories to integer codes.
This is simple but imposes an ordinal relationship. For non-ordinal categories, one-hot encoding is often preferred.
Here we demonstrate label encoding for variety.
First convert to ‘category’ dtype, then use
.cat.codesto get integer codes.Note: In practice, use sklearn’s LabelEncoder or OrdinalEncoder for more control.
# Convert admission_unit to 'category' dtype
patients_cleaned['admission_unit_encoded'] = patients_cleaned['admission_unit'].astype('category')
# Label encode admission_unit
patients_cleaned['admission_unit_encoded'] = patients_cleaned['admission_unit_encoded'].cat.codes
patients_cleaned.head()
| patient_id | age | admission_date | discharge_date | weight | height | blood_pressure | admission_unit | albumin_g_dl | gender_male | admission_unit_encoded | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 39.0 | 2023-01-05 | 2023-01-10 | 78.0 | 184.0 | 130.0 | Surgery | 1.00 | False | 9 |
| 1 | 2 | 60.0 | 2023-02-11 | 2023-02-20 | 55.0 | 180.0 | 90.0 | Cardiology | 4.26 | True | 0 |
| 2 | 4 | 50.0 | 2023-03-20 | 2023-03-25 | 70.0 | 179.0 | 110.0 | ICU | 3.10 | False | 3 |
| 3 | 10 | 45.0 | 2023-07-05 | 2023-07-15 | 90.0 | 165.0 | 80.0 | Neurology | 3.10 | True | 4 |
| 4 | 12 | 90.0 | 2023-08-10 | 2023-08-20 | 45.0 | 170.0 | 60.0 | ICU | 4.26 | False | 3 |
3 Feature engineering - creating new features from existing ones#
BMI (Body Mass Index) is a common clinical feature derived from weight and height.
BMI = weight (kg) / (height (m))2
In our dataset height is in cm, so it needs to be converted before calculation.
# Compute BMI (height in cm -> convert to meters)
patients_cleaned['BMI'] = patients_cleaned['weight'] / (patients_cleaned['height']/100)**2
# Inspect BMI distribution and missingness
patients_cleaned['BMI'].describe()
count 26.000000
mean 25.087716
std 6.945768
min 14.527376
25% 21.232993
50% 22.505044
75% 29.602852
max 40.562466
Name: BMI, dtype: float64
Save transformed data#
# Save the transformed dataset
patients_cleaned.to_csv("data/patients_transformed.csv", index=False)
4 Scaling and standardization#
Many ML algorithms assume features are on similar scales
MinMaxScalerrescales to [0,1]StandardScalercenters to mean=0 and std=1
We demonstrate both for different use cases.
Method 1: Min-Max Scaling#
# Demonstrate MinMax scaling for 'age' and Standard scaling for blood pressure
minmax = MinMaxScaler()
patients_cleaned['age_minmax'] = minmax.fit_transform(patients_cleaned[['age']])
Method 2: Standardization#
std = StandardScaler()
patients_cleaned['bp_std'] = std.fit_transform(patients_cleaned[['blood_pressure']])
patients_cleaned.head()
| patient_id | age | admission_date | discharge_date | weight | height | blood_pressure | admission_unit | albumin_g_dl | gender_male | admission_unit_encoded | BMI | age_minmax | bp_std | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 39.0 | 2023-01-05 | 2023-01-10 | 78.0 | 184.0 | 130.0 | Surgery | 1.00 | False | 9 | 23.038752 | 0.105263 | 1.274066 |
| 1 | 2 | 60.0 | 2023-02-11 | 2023-02-20 | 55.0 | 180.0 | 90.0 | Cardiology | 4.26 | True | 0 | 16.975309 | 0.473684 | -0.436090 |
| 2 | 4 | 50.0 | 2023-03-20 | 2023-03-25 | 70.0 | 179.0 | 110.0 | ICU | 3.10 | False | 3 | 21.847009 | 0.298246 | 0.418988 |
| 3 | 10 | 45.0 | 2023-07-05 | 2023-07-15 | 90.0 | 165.0 | 80.0 | Neurology | 3.10 | True | 4 | 33.057851 | 0.210526 | -0.863629 |
| 4 | 12 | 90.0 | 2023-08-10 | 2023-08-20 | 45.0 | 170.0 | 60.0 | ICU | 4.26 | False | 3 | 15.570934 | 1.000000 | -1.718707 |
Further preprocessing for modeling#
scaling all numeric features
dropping any remaining irrelevant columns
handling any remaining missing values
drop identifier
patient_id
Exercise — Data Transformation#
Standardize the
blood_pressurecolumn (use StandardScaler - is already imported), store asbp_standardized.Create a new feature
length_of_stayas the difference in days betweendischarge_dateandadmission_date. (Hint: columns need to be datetime dtype)Plot a histogram of the computed BMI.
# 1. Standardize blood_pressure
# 2. Create length_of_stay feature
# 3. Plot histogram of BMI
