Generating images using DALL-E#
In this notebook we will demonstrate how to ask openAI’s DALL-E model to generate some images. Depending on what we request and how the model was trained, the results may be more or less appropriate. We will again use a small helper function to return the image directly.
Read more:
import openai
from skimage.io import imread, imshow
from numpy import random
from matplotlib import pyplot as plt
def prompt_image(message:str, width:int=1024, height:int=1024, model='dall-e-3'):
client = openai.OpenAI()
response = client.images.generate(
prompt=message,
model=model,
n=1,
size=f"{width}x{height}"
)
image_url = response.data[0].url
image = imread(image_url)
return image
When generating scientific images, they may, or may not be a proper representation. Presumably, DALL-E was trained on many natural images (showing cars, trees, houses, etc.) and not so many images of cells.
nuclei = prompt_image('an image of 100 nuclei in a fluorescence microscopy image')
imshow(nuclei)
<matplotlib.image.AxesImage at 0x292557e9890>
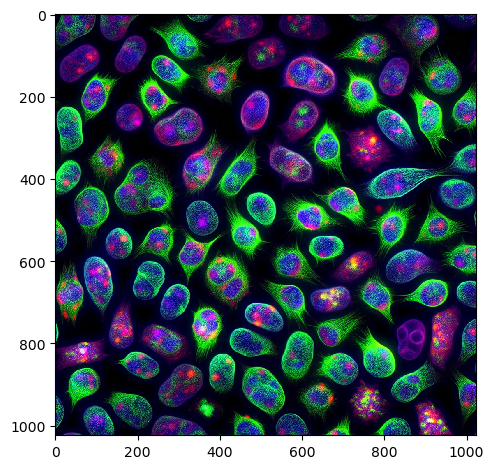
Maybe, generating images of histological slices is more promising.
histology = prompt_image('a histology image of lung cancer cells and some healthy tissue')
imshow(histology)
<matplotlib.image.AxesImage at 0x292558a0790>
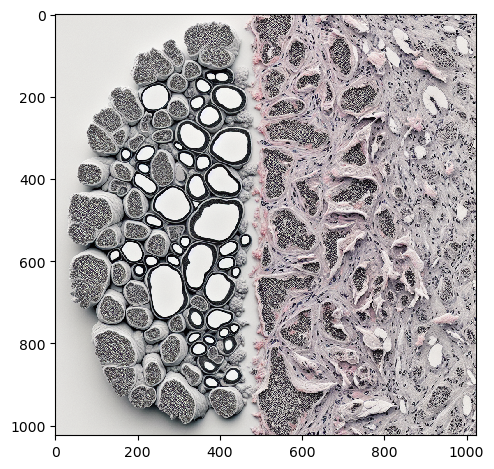
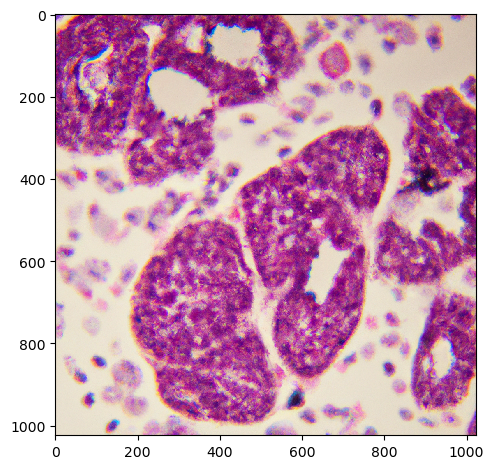
In some cases, Dall-E 2 produces more realistic output.
histology = prompt_image('a histology image of lung cancer cells and some healthy tissue',
model='dall-e-2')
imshow(histology)
<matplotlib.image.AxesImage at 0x29255e3ef90>
Exercise#
Try to generate realistically looking electron microscopy images. Try Dall-E 2 and 3. How about fluorescence microscopy images? How about CT and MRI?
