Handling missing data#
When analyzing tabular data, sometimes table cells are present that do not contain data. In Python, this typically means the value is Not a Number, or NaN. We need to deal with these NaN entries differently, and this notebook will introduce how.
To get a first view where NaNs play a role, we load again some example data from a CSV file. This file uses a semicolon as separator or delimiter…we can provide pandas with this information.
import pandas as pd
import matplotlib.pyplot as plt
# Now that we know about our datatypes, we can provide this info when loading the DataFrame
# - parse timestamp to datetime and set as index
# - parse label to string, valid to boolean
df = pd.read_csv('data/converted_measuremets.csv', index_col=0, parse_dates=True, dtype={'label': 'string', 'valid': 'boolean'})
df.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 11 entries, 2023-01-01 07:00:00 to 2023-01-01 20:00:00
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 label 11 non-null string
1 valid 10 non-null boolean
2 value1 10 non-null float64
3 value2 8 non-null float64
4 value3 9 non-null float64
5 value4 10 non-null float64
6 value5 8 non-null float64
dtypes: boolean(1), float64(5), string(1)
memory usage: 638.0 bytes
df
| label | valid | value1 | value2 | value3 | value4 | value5 | |
|---|---|---|---|---|---|---|---|
| timestamp | |||||||
| 2023-01-01 07:00:00 | A | True | 9.0 | 2.0 | 3.1 | 0.98 | 1.23 |
| 2023-01-01 08:00:00 | B | False | 11.0 | NaN | 0.4 | 0.40 | 1.11 |
| 2023-01-01 10:00:00 | D | True | 9.5 | 5.0 | NaN | 2.56 | NaN |
| 2023-01-01 10:15:00 | C | True | 10.2 | 8.0 | 5.6 | NaN | NaN |
| 2023-01-01 11:00:00 | E | False | 15.0 | 7.0 | 4.4 | 3.14 | 2.34 |
| 2023-01-01 12:00:00 | F | True | 400.0 | NaN | 2.2 | 2.71 | 2.45 |
| 2023-01-01 16:00:00 | G | False | 9.0 | 1.0 | 1.1 | 3.58 | 0.98 |
| 2023-01-01 17:00:00 | H | True | 11.0 | 4.0 | 0.9 | 4.12 | 1.05 |
| 2023-01-01 18:00:00 | I | True | 11.3 | 6.0 | 3.3 | 3.33 | 1.67 |
| 2023-01-01 19:00:00 | J | False | 10.0 | 9.0 | 4.1 | 2.98 | 2.89 |
| 2023-01-01 20:00:00 | K | <NA> | NaN | NaN | NaN | 3.87 | NaN |
Examine missing data#
As you can see, there are rows containing NaNs. A check if there are NaNs anywhere in a DataFrame is an important quality check at the beginning of any analysis task and good scientific practice.
Pandas’ isnull provides a boolean masking of the DataFrame with True / False values, where True indicates a NaN.
df.isnull()
| label | valid | value1 | value2 | value3 | value4 | value5 | |
|---|---|---|---|---|---|---|---|
| timestamp | |||||||
| 2023-01-01 07:00:00 | False | False | False | False | False | False | False |
| 2023-01-01 08:00:00 | False | False | False | True | False | False | False |
| 2023-01-01 10:00:00 | False | False | False | False | True | False | True |
| 2023-01-01 10:15:00 | False | False | False | False | False | True | True |
| 2023-01-01 11:00:00 | False | False | False | False | False | False | False |
| 2023-01-01 12:00:00 | False | False | False | True | False | False | False |
| 2023-01-01 16:00:00 | False | False | False | False | False | False | False |
| 2023-01-01 17:00:00 | False | False | False | False | False | False | False |
| 2023-01-01 18:00:00 | False | False | False | False | False | False | False |
| 2023-01-01 19:00:00 | False | False | False | False | False | False | False |
| 2023-01-01 20:00:00 | False | True | True | True | True | False | True |
With this boolean masking, we can do some further analysis. And since True / False can also be interpreted as 1 / 0, we also can do math with it.
# Check if there are any NaN in the data
df.isna().values.any()
np.True_
# Get a column-wise overview of NaN and count them
df.isnull().sum().sort_values(ascending=False)
value2 3
value5 3
value3 2
valid 1
value1 1
value4 1
label 0
dtype: int64
# Compute the column-wise percentage of NaN
df.isnull().mean().sort_values(ascending=False) * 100
value2 27.272727
value5 27.272727
value3 18.181818
valid 9.090909
value1 9.090909
value4 9.090909
label 0.000000
dtype: float64
For most DataFrame methods, we can provide the parameter axis, determining whether the computation should be done on the columns or the rows / index.
# Compute the row-wise percentage of NaN
df.isnull().mean(axis=1).sort_values(ascending=False) * 100
timestamp
2023-01-01 20:00:00 71.428571
2023-01-01 10:00:00 28.571429
2023-01-01 10:15:00 28.571429
2023-01-01 08:00:00 14.285714
2023-01-01 12:00:00 14.285714
2023-01-01 07:00:00 0.000000
2023-01-01 11:00:00 0.000000
2023-01-01 16:00:00 0.000000
2023-01-01 17:00:00 0.000000
2023-01-01 18:00:00 0.000000
2023-01-01 19:00:00 0.000000
dtype: float64
We may also want to the subset of rows containing NaNs. Here, we can combine the indexing with a boolean masking for existing NaN:
df.loc[df.isnull().any(axis=1)]
| label | valid | value1 | value2 | value3 | value4 | value5 | |
|---|---|---|---|---|---|---|---|
| timestamp | |||||||
| 2023-01-01 08:00:00 | B | False | 11.0 | NaN | 0.4 | 0.40 | 1.11 |
| 2023-01-01 10:00:00 | D | True | 9.5 | 5.0 | NaN | 2.56 | NaN |
| 2023-01-01 10:15:00 | C | True | 10.2 | 8.0 | 5.6 | NaN | NaN |
| 2023-01-01 12:00:00 | F | True | 400.0 | NaN | 2.2 | 2.71 | 2.45 |
| 2023-01-01 20:00:00 | K | <NA> | NaN | NaN | NaN | 3.87 | NaN |
Handling NaNs - Drop data#
Depending on what kind of data analysis should be performed, it might make sense to just ignore rows and columns that contain NaN values. Alternatively, it is possible to delete rows or columns that contain NaNs. We may also use different methods to impute missing data and fill the gaps, where we should consider that those values may not represent the “real world” and may have an impact on further data analysis.
It always depends on your project, the data, and what is important or not for your analysis. It’s not an easy answer. Whatever the solution will be, it should be documented and this info should also be provided in any scientific publication based on this data.
In this case, we’ll use dropna() to remove all rows (index) which have any missing data.
df_no_nan = df.dropna(axis='index', how='any')
df_no_nan.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 6 entries, 2023-01-01 07:00:00 to 2023-01-01 19:00:00
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 label 6 non-null string
1 valid 6 non-null boolean
2 value1 6 non-null float64
3 value2 6 non-null float64
4 value3 6 non-null float64
5 value4 6 non-null float64
6 value5 6 non-null float64
dtypes: boolean(1), float64(5), string(1)
memory usage: 348.0 bytes
We can now also check again if NaNs are present.
df_no_nan.isnull().values.any()
np.False_
Exercise#
Instead of removing rows with any missing data, remove all rows that contain more than 50% of NaN. Refer to the docs for dopna() for help.
Handling NaNs - Imputation#
Instead of dropping incomplete data, we can try to impute data to fill the gaps. Pandas provides basic methods to fill missing data, while other libraries also provide more sophisticated approaches, e.g.:
# Fill missing numerical values with the mean of the column
df.fillna(df.select_dtypes(include='number').mean())
| label | valid | value1 | value2 | value3 | value4 | value5 | |
|---|---|---|---|---|---|---|---|
| timestamp | |||||||
| 2023-01-01 07:00:00 | A | True | 9.0 | 2.00 | 3.100000 | 0.980 | 1.230 |
| 2023-01-01 08:00:00 | B | False | 11.0 | 5.25 | 0.400000 | 0.400 | 1.110 |
| 2023-01-01 10:00:00 | D | True | 9.5 | 5.00 | 2.788889 | 2.560 | 1.715 |
| 2023-01-01 10:15:00 | C | True | 10.2 | 8.00 | 5.600000 | 2.767 | 1.715 |
| 2023-01-01 11:00:00 | E | False | 15.0 | 7.00 | 4.400000 | 3.140 | 2.340 |
| 2023-01-01 12:00:00 | F | True | 400.0 | 5.25 | 2.200000 | 2.710 | 2.450 |
| 2023-01-01 16:00:00 | G | False | 9.0 | 1.00 | 1.100000 | 3.580 | 0.980 |
| 2023-01-01 17:00:00 | H | True | 11.0 | 4.00 | 0.900000 | 4.120 | 1.050 |
| 2023-01-01 18:00:00 | I | True | 11.3 | 6.00 | 3.300000 | 3.330 | 1.670 |
| 2023-01-01 19:00:00 | J | False | 10.0 | 9.00 | 4.100000 | 2.980 | 2.890 |
| 2023-01-01 20:00:00 | K | <NA> | 49.6 | 5.25 | 2.788889 | 3.870 | 1.715 |
# Fill missing boolean values with the most frequent one
df.fillna(df.select_dtypes(include='boolean').mode().iloc[0])
| label | valid | value1 | value2 | value3 | value4 | value5 | |
|---|---|---|---|---|---|---|---|
| timestamp | |||||||
| 2023-01-01 07:00:00 | A | True | 9.0 | 2.0 | 3.1 | 0.98 | 1.23 |
| 2023-01-01 08:00:00 | B | False | 11.0 | NaN | 0.4 | 0.40 | 1.11 |
| 2023-01-01 10:00:00 | D | True | 9.5 | 5.0 | NaN | 2.56 | NaN |
| 2023-01-01 10:15:00 | C | True | 10.2 | 8.0 | 5.6 | NaN | NaN |
| 2023-01-01 11:00:00 | E | False | 15.0 | 7.0 | 4.4 | 3.14 | 2.34 |
| 2023-01-01 12:00:00 | F | True | 400.0 | NaN | 2.2 | 2.71 | 2.45 |
| 2023-01-01 16:00:00 | G | False | 9.0 | 1.0 | 1.1 | 3.58 | 0.98 |
| 2023-01-01 17:00:00 | H | True | 11.0 | 4.0 | 0.9 | 4.12 | 1.05 |
| 2023-01-01 18:00:00 | I | True | 11.3 | 6.0 | 3.3 | 3.33 | 1.67 |
| 2023-01-01 19:00:00 | J | False | 10.0 | 9.0 | 4.1 | 2.98 | 2.89 |
| 2023-01-01 20:00:00 | K | True | NaN | NaN | NaN | 3.87 | NaN |
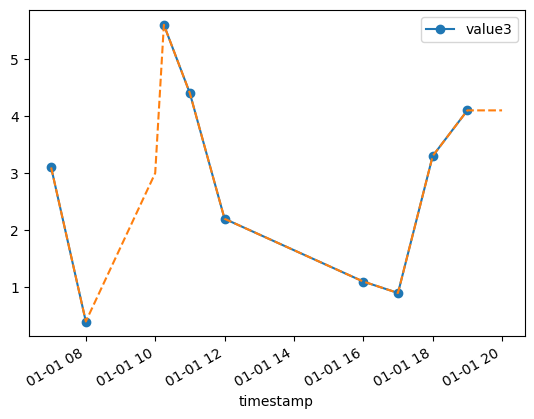
# We can also try to interpolate missing values
df['value3'].plot(style="o-", legend=True)
# Interpolate privides different methods, e.g linear, quadratic, etc
df['value3'].interpolate(method='linear').plot(style="--")
plt.show()
Exercise#
Lets combine all this to
fill missing boolean values via mode
fill missing numerical values by linear interpolation
You may want to consider the use of apply(), to apply according methods to each column.
Save the result to data/filled_measurements.csv
