Tiling images with overlap#
When processing images in tiles, we can observe artifacts on borders of the tiles in the resulting image. One strategy to prevent these artifacts is to process tiles with some overlap. dask also supports this. Again, for demonstration purposes, we use imshow to show the resulting images. If these were big data, the imshow function would not work.
import dask
import dask.array as da
from skimage.filters import gaussian
from skimage.data import cells3d
from stackview import imshow
from skimage.io import imread
Similar to the example in the last lesson, we define a procedure that applies a Gaussian blur to an image and prints out the size of the image, just so that we know:
def procedure(image):
print("processing", image.shape)
return gaussian(image, sigma=5)
image = imread("data/nuclei2d.tif")
imshow(image)
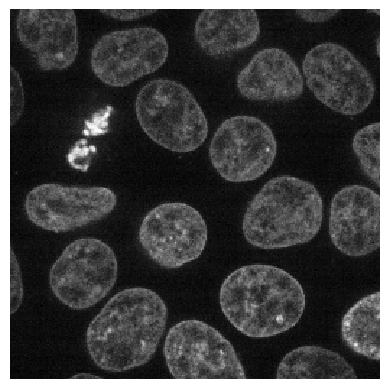
After loading the image, we tile it as usual.
tiles = da.from_array(image, chunks=(128, 128))
tiles
|
||||||||||||||||
Next, we tell dask what to do with our tiles: We want to map the function procedure to the tiles with a defined overlap.
overlap_width = 1
tile_map = da.map_overlap(procedure, tiles, depth=overlap_width)
processing (0, 0)
processing (1, 1)
The function was executed twice with very small images (0x0 and 1x1 pixels) to check if it works. Next, we actually compute the result.
result = tile_map.compute() # Warning: This loads all image data into memory
processing (129, 129)
processing (129, 129)
processing (129, 129)
processing (129, 129)
processing (128, 128)
processing (128, 128)
processing (128, 128)
processing (128, 128)
processing (132, 132)
processing (132, 132)
processing (140, 140)
processing (140, 140)
processing (140, 140)
processing (144, 144)
processing (144, 144)
From the printed image size, we can see that the processed image size is 2 pixels larger than the tile size. That’s the overlap of 1 pixel in all directions.
Minimizing border effects#
Next, we will compare the result when processing the whole image with the image processed in tiles with different overlaps. This gives us the chance to figure out the minimum necessary overlap width for eliminating border effects. First, we compute the result for the full image.
untiled_result = procedure(image)
processing (256, 256)
Then, we run a for-loop with different border_widths.
for overlap_width in range(0, 10, 2):
print("Overlap width", overlap_width)
tile_map = da.map_overlap(procedure, tiles, depth=overlap_width, boundary='nearest')
result = tile_map.compute()
difference = result - untiled_result
imshow(difference)
print("sum difference", difference.sum())
print("-----------------------------------")
Overlap width 0
processing (0, 0)
processing (1, 1)

sum difference 1.528818863147824
-----------------------------------
Overlap width 2
processing (0, 0)
processing (1, 1)
processing (132, 132)
processing (132, 132)

sum difference 5.971294396376637
-----------------------------------
Overlap width 4
processing (0, 0)
processing (1, 1)
processing (136, 136)
processing (136, 136)
processing (136, 136)
processing (136, 136)
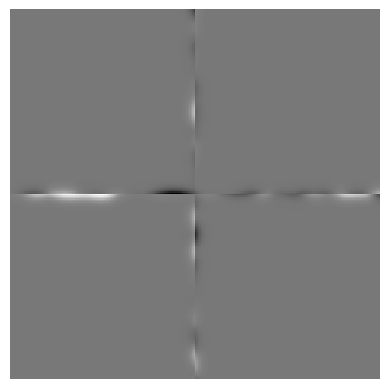
sum difference 3.823608171106622
-----------------------------------
Overlap width 6
processing (0, 0)
processing (1, 1)
processing (140, 140)

sum difference 0.9650985532492449
-----------------------------------
Overlap width 8
processing (0, 0)
processing (1, 1)
processing (144, 144)
processing (144, 144)

sum difference 0.3836489788163834
-----------------------------------
As you can see, the sum difference becomes smaller the wider the overlap is. This is obviously related to the procedure we applied. Depending on the filters applied to the image, we need a different overlap width to eliminate the error.
Exercise#
Determine the overlap width you need to get artifact-free images.
