VLMs guessing image segmentation strategies#
In this notebook we present images to VLMs and ask them which algorithm to use for segmenting the image. One could expect that depending on the image, the VLM suggests different strategies. In a second example, we demonstrate how a list of rules can be used to guide the VLM in guiding us.
import openai
import PIL
import stackview
from skimage import data
from skimage.io import imread
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import wordcloud
We will need some helper functions for assembling a prompt and submitting it to the openai server.
def prompt_with_image(message:str, image=None, model="gpt-4o-2024-11-20"):
"""A prompt helper function that sends a text message and an image
to openAI and returns the text response.
"""
import os
# convert message in the right format if necessary
if isinstance(message, str):
message = [{"role": "user", "content": message}]
if image is not None:
image_message = image_to_message(image)
else:
image_message = []
# setup connection to the LLM
client = openai.OpenAI()
# submit prompt
response = client.chat.completions.create(
model=model,
messages=message + image_message
)
# extract answer
return response.choices[0].message.content
def image_to_message(image):
import base64
from stackview._image_widget import _img_to_rgb
rgb_image = _img_to_rgb(image)
byte_stream = numpy_to_bytestream(rgb_image)
base64_image = base64.b64encode(byte_stream).decode('utf-8')
return [{"role": "user", "content": [{
"type": "image_url",
"image_url": {
"url": f"data:image/jpeg;base64,{base64_image}"
}
}]}]
def numpy_to_bytestream(data):
"""Turn a NumPy array into a bytestream"""
import numpy as np
from PIL import Image
import io
# Convert the NumPy array to a PIL Image
image = Image.fromarray(data.astype(np.uint8)).convert("RGBA")
# Create a BytesIO object
bytes_io = io.BytesIO()
# Save the PIL image to the BytesIO object as a PNG
image.save(bytes_io, format='PNG')
# return the beginning of the file as a bytestream
bytes_io.seek(0)
return bytes_io.read()
These are the example images we will be using.
membrane_image = data.cells3d()[30, 0]
stackview.insight(membrane_image)
|
|
|
nuclei_image = data.human_mitosis()[100:200,:100]
stackview.insight(nuclei_image)
|
|
|
dense_nuclei_image = data.human_mitosis()[330:430, 355:455]
stackview.insight(dense_nuclei_image)
|
|
|
hela_cells = imread("data/hela-cells-8bit.tif")
stackview.insight(hela_cells)
|
|
|
This helper function will send the image together with a prompt to the LLM service provider and display a word cloud of the suggested algorithms.
def determine_algorithm(prompt, image, num_samples=25):
responses = []
for _ in range(num_samples):
responses.append(prompt_with_image(prompt, image))
responses = [r.lower().replace("algorithm", "").replace("segmentation", "").strip() for r in responses]
text = " ".join(responses)
# Generate word cloud
w = wordcloud.WordCloud(width=800, height=400, colormap='viridis', background_color='white').generate(text)
# Display the word cloud
plt.imshow(w, interpolation='bilinear')
plt.show()
First, we prompt for microscopy image segmentation algorithms with no details and no image.
prompt = """You are a bioimage-analysis expert.
What is the best image processing algorithm to segment microscopy images?
Answer the algorithm name only. No explanations.
"""
determine_algorithm(prompt, None)
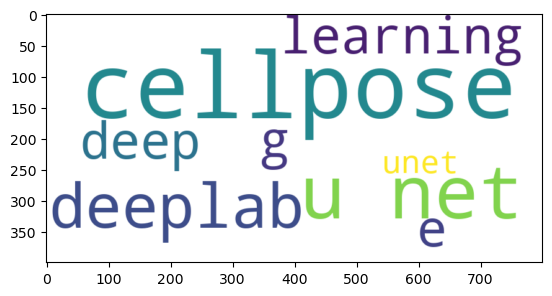
This is the simple prompt we submit to the server.
prompt = """You are a bioimage-analysis expert.
What is the best image processing algorithm to segment this microscopy image?
Answer the algorithm name only. No explanations.
"""
determine_algorithm(prompt, membrane_image)

determine_algorithm(prompt, nuclei_image)

determine_algorithm(prompt, dense_nuclei_image)

determine_algorithm(prompt, hela_cells)

Next, we try the same strategy using a more complex prompt containing a list of rules to guide the VLM.
prompt = """You are a bioimage-analysis expert. You have a rule-book what alogrithms to use for specific images.
## Rules
* If an image shows sparse objects such as nuclei, use Otsu-thresholding for segmenting them.
* If an image shows dense, partially overlapping objects such as nuclei, use StarDist.
* If an image shows large cell-like structures with bright membranes, use the Watershed algorithm.
* In case of doubt, use CellPose.
## The task
What is the best image processing algorithm to segment this microscopy image?
Answer the algorithm name only. No explanations.
"""
determine_algorithm(prompt, membrane_image)

determine_algorithm(prompt, nuclei_image)

determine_algorithm(prompt, dense_nuclei_image)

determine_algorithm(prompt, hela_cells)

Exercise#
Load a natural picture, e.g. showing of a cat, and ask the LLM how to process the image using both prompts above.
