Cropping images#
When working with microscopy images, it often makes limited sense to process the whole image. We typically crop out interesting regions and process them in detail.
from skimage.io import imread, imshow
#from microfilm.microplot import microshow
image = imread("data/blobs.tif")
Before we can crop an image, we may want to know its precise shape (dimensions):
image.shape
(254, 256)
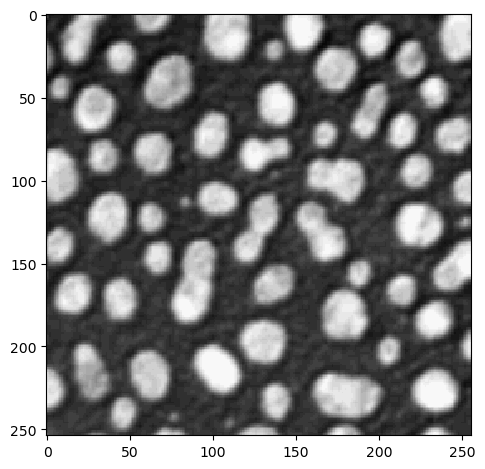
Recap: Visualization using imshow:
imshow(image);
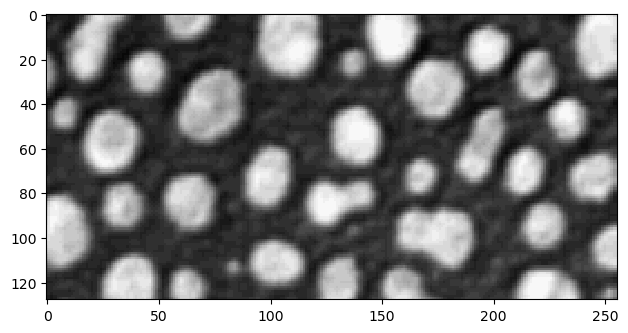
Cropping images works exactly like cropping lists and tuples, by using indices to specify the range of elements to use:
cropped_image1 = image[0:128]
imshow(cropped_image1);
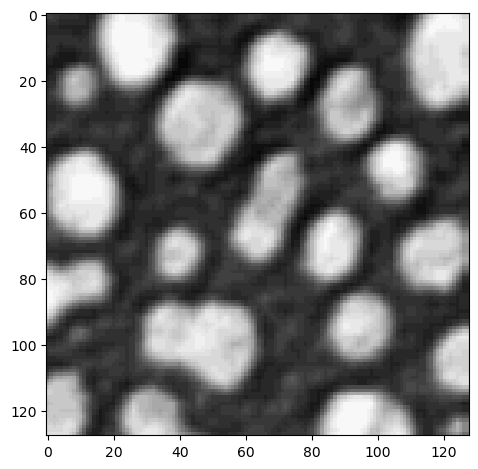
To crop the image in the second dimension as well, we add a , in the square brackets:
cropped_image2 = image[0:128, 128:]
imshow(cropped_image2);
Sub-sampling images#
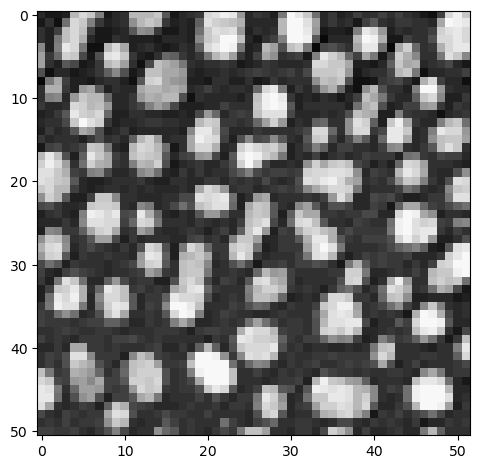
Also step sizes can be specified as if we would process lists and tuples. Technically, we are sub-sampling the image in this case. We sample a subset of the original pixels for example in steps of 5:
sampled_image = image[::5, ::5]
imshow(sampled_image);
Flipping images#
Negative step sizes flip the image.
flipped_image = image[::, ::-1]
imshow(flipped_image);
Exercise#
Open the banana020.tif data set and crop out the region where the banana slice is located.
