The [seeded] watershed algorithm#
The watershed algorithm is traditionally used for cell segmentation in microscopy images when a membrane marker was used. If image quality and resolution is high, the algorithm works great. In cases where high image quality cannot be achieved, deep-learning approaches are outperforming the watershed algorithm.
We will use the scriptable napari plugin napari-segment-blobs-and-things-with-membranes. Under the hood, this plugin uses functions from scikit-image. We also use napari-simpleitk-image-processing which is based on the Insight Toolkit, maybe the oldest open-source image processing library on earth.
import napari_segment_blobs_and_things_with_membranes as nsbatwm
import napari_simpleitk_image_processing as nsitk
from skimage.io import imread, imshow
from skimage.data import cells3d
import stackview
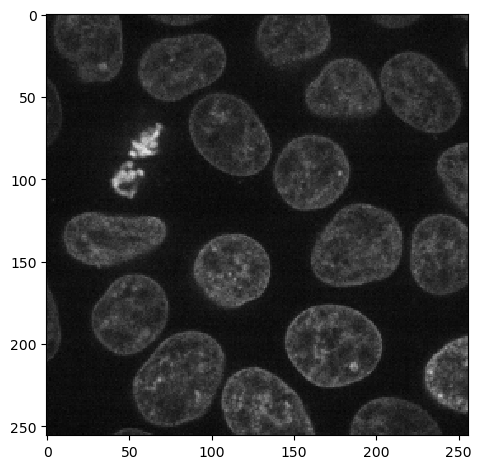
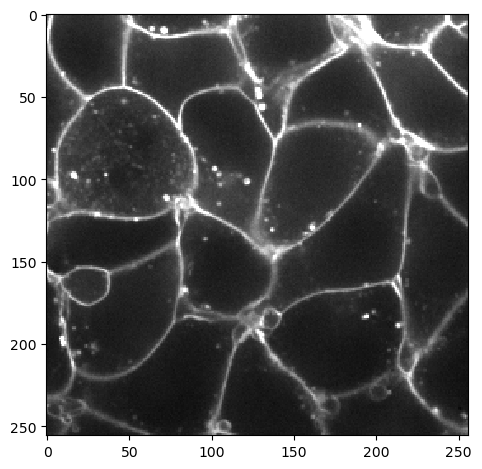
As example we will use a single 2d slice from the cells3d dataset from scikit-image. The image has two channels: A nuclei-channel and a channel showing cell membranes.
image_stack = cells3d()
nuclei_image = image_stack[30, 1]
imshow(nuclei_image)
<matplotlib.image.AxesImage at 0x16f1e19a0a0>
membrane_image = image_stack[30, 0]
imshow(membrane_image, vmax=10000)
<matplotlib.image.AxesImage at 0x16f1f1c37f0>
Before we can apply the seeded watershed algorithm, we need seeds. The nuclei channel allows us to make seeds, e.g. using the Voronoi-Otsu-Labeling algorithm.
nuclei_labels = nsbatwm.voronoi_otsu_labeling(nuclei_image, spot_sigma=10, outline_sigma=1)
nuclei_labels
|
|
nsbatwm made image
|
We can then use these seeds to flood the membrane-image using the watershed algorithm.
cell_labels = nsbatwm.seeded_watershed(membrane_image, nuclei_labels)
cell_labels
|
|
nsbatwm made image
|
In case the segmentation would not look perfect, it is common to apply pre-processing filters to the membrane image. This allows to make outlines smoother for example:
blurred_image = nsbatwm.gaussian_blur(membrane_image, sigma=4)
blurred_image
|
|
nsbatwm made image
|
cell_labels2 = nsbatwm.seeded_watershed(blurred_image, nuclei_labels)
cell_labels2
|
|
nsbatwm made image
|
Splitting touching objects#
There are other use-cases for the watershed algorithm. In this section we will split objects in binary images that have a roundish shape and touch each other.
Starting point for this is a binary image, e.g. made using thresholding.
blobs = imread('../03b_image_processing/data/blobs.tif')
stackview.insight(blobs)
|
|
|
binary = nsbatwm.threshold_otsu(blobs).astype(bool)
binary
<__array_function__ internals>:180: RuntimeWarning: Converting input from bool to <class 'numpy.uint8'> for compatibility.
|
|
nsbatwm made image
|
We can then split the touching object by only taking the binary image into account. The underlying algorithm aims to produce similar results to ImageJ’s binary watershed algorithm and the implementation here also works in 3D.
split_objects = nsbatwm.split_touching_objects(binary)
split_objects
<__array_function__ internals>:180: RuntimeWarning: Converting input from bool to <class 'numpy.uint8'> for compatibility.
|
|
nsbatwm made image
|
It is also possible to retrieve a label image as result. Note that in this case, the black line/gap between objects will not be present.
touching_labels = nsitk.touching_objects_labeling(binary)
touching_labels
|
|
n-sitk made image
|
