Morphological Image Processing#
Morphological operations transform images based on shape; typically we mean binary images in this context.
import numpy as np
from skimage.io import imread
import matplotlib.pyplot as plt
from skimage import morphology
from skimage import filters
Kernels, footsprints and structural elements#
If we work with scikit-image, many morphological filters have a footprint parameter. This footprint is the filter kernel, and in the literature you also find the term structural element for this.
disk = morphology.disk(3) # creates a disk of 1 with radius = 3
disk
array([[0, 0, 0, 1, 0, 0, 0],
[0, 1, 1, 1, 1, 1, 0],
[0, 1, 1, 1, 1, 1, 0],
[1, 1, 1, 1, 1, 1, 1],
[0, 1, 1, 1, 1, 1, 0],
[0, 1, 1, 1, 1, 1, 0],
[0, 0, 0, 1, 0, 0, 0]], dtype=uint8)
plt.imshow(disk, cmap='gray')
<matplotlib.image.AxesImage at 0x7fb952653dc0>
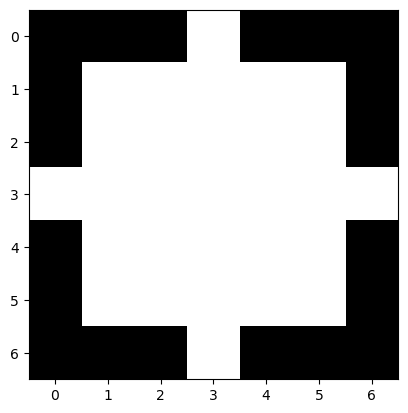
square = morphology.square(3) # creates a square with widht and height = 3
square
array([[1, 1, 1],
[1, 1, 1],
[1, 1, 1]], dtype=uint8)
Binary morphology#
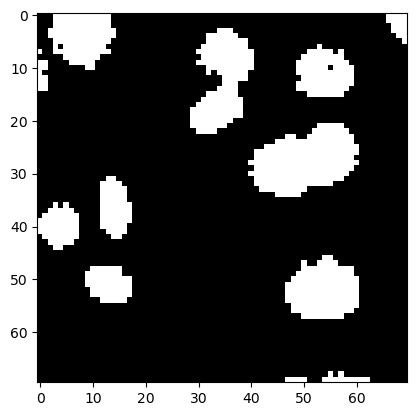
For demonstrating morphological filtering of binary images, we use the small nuclei image again.
image_nuclei = imread('data/mitosis_mod.tif').astype(float)
image_binary = image_nuclei > filters.threshold_otsu(image_nuclei)
plt.imshow(image_binary, cmap='gray')
<matplotlib.image.AxesImage at 0x7fb9523edb50>
Erosion and Dilation#
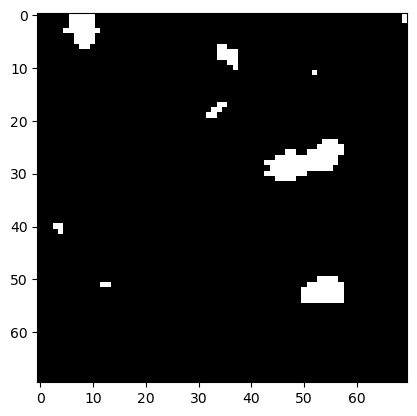
To make white islands in the black ocean smaller, we need to erode its coastlines.
eroded = morphology.binary_erosion(image_binary, disk)
plt.imshow(eroded, cmap='gray')
<matplotlib.image.AxesImage at 0x7fb952352880>
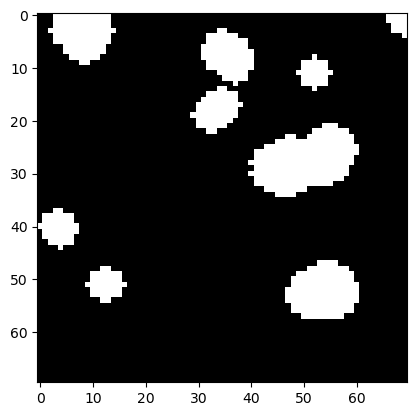
If we dilate the image afterwards, we get white islands back that look smoother than in the original binary image.
eroded_dilated = morphology.binary_dilation(eroded, disk)
plt.imshow(eroded_dilated, cmap='gray')
<matplotlib.image.AxesImage at 0x7fb952360310>
Calling erosion and dilation subsequently is so common that there is an extra function which does exactly that. As the gap between islands open the operation is called opening.
opened = morphology.binary_opening(image_binary, disk)
plt.imshow(opened, cmap='gray')
<matplotlib.image.AxesImage at 0x7fb952265520>

Exercise 1#
There is also a closing operation. Apply it to the binary image.
Exercise 2#
Search the scikit-image documentation for minimum and maximum filters. Apply the minimum filter to the binary image and the maximum filter to the result afterwards. Compare it to the images shown above.
Hint: You need to covert your binary image (array of true/false) to an integer array (0/1), therfore you can use the numpy function np.uint8.
