Segment DAPI channel - batch process a folder of images#
from skimage.io import imread
import pyclesperanto_prototype as cle # version 0.24.3
import napari_segment_blobs_and_things_with_membranes as nsbatwm # version 0.3.8
from skimage import morphology
from skimage.morphology import disk, binary_erosion
import napari
import numpy as np
from tqdm import tqdm
import matplotlib.pyplot as plt
import os
import tifffile as tif
# create napari viewer instance
if 'viewer' not in globals():
viewer = napari.Viewer()
path = "/Users/laura/projects/Bio-image_analysis_school_ScadsAI/prepared_dataset"
prepared_images = os.listdir(path)
prepared_images
['aphidicolin_timelapse.tif',
'.DS_Store',
'nocodazole_timelapse.tif',
'AZ-I_timelapse.tif',
'taxol_timelapse.tif',
'labels_nuclei',
'latrunculin B_timelapse.tif',
'epothilone B_timelapse.tif',
'monastrol_timelapse.tif',
'AZ-H_timelapse.tif',
'DMSO_timelapse.tif',
'colchicine_timelapse.tif',
'doxorubicin_timelapse.tif',
'cytochalasin B_timelapse.tif',
'labels_tubulin',
'labels_actin',
'AZ-A_timelapse.tif']
Process one image as an example#
filename = 'latrunculin B_timelapse'
img = tif.imread(os.path.join(path, f'{filename}.tif'))
# viewer.add_image(img) # and then in the viewer right click on the layer - split RGB or:
viewer.add_image(
img,
name=["tubulin", "actin", "nuclei"],
colormap=["magenta", "green", "blue"],
channel_axis=3)
[<Image layer 'tubulin' at 0x16e581670>,
<Image layer 'actin' at 0x16e5816a0>,
<Image layer 'nuclei' at 0x176ebfc40>]
image = viewer.layers['nuclei'].data
labels = []
for t in tqdm(range(image.shape[0])):
# gaussian blur
blurred_image = cle.gaussian_blur(image[t], None, 2.0, 2.0, 0.0)
bg_removed = cle.top_hat_sphere(blurred_image, None, 20.0, 20.0, 0.0)
# threshold otsu
binary_nuclei = cle.threshold_otsu(bg_removed)
# split touching objects
binary_nuclei_edited = nsbatwm.split_touching_objects(binary_nuclei, 4.0)
# additional erosion to split a few still touching nuclei
binary_eroded_nuclei = binary_erosion(binary_nuclei_edited, disk(2))
# connected component labeling
labelled_nuclei = nsbatwm.connected_component_labeling(binary_eroded_nuclei, False)
# expand eroded nuclei back
dilated_labels = morphology.dilation(labelled_nuclei, disk(2))
labels.append(dilated_labels)
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 8/8 [00:03<00:00, 2.29it/s]
labels_timelapse = np.stack(labels, axis=0)
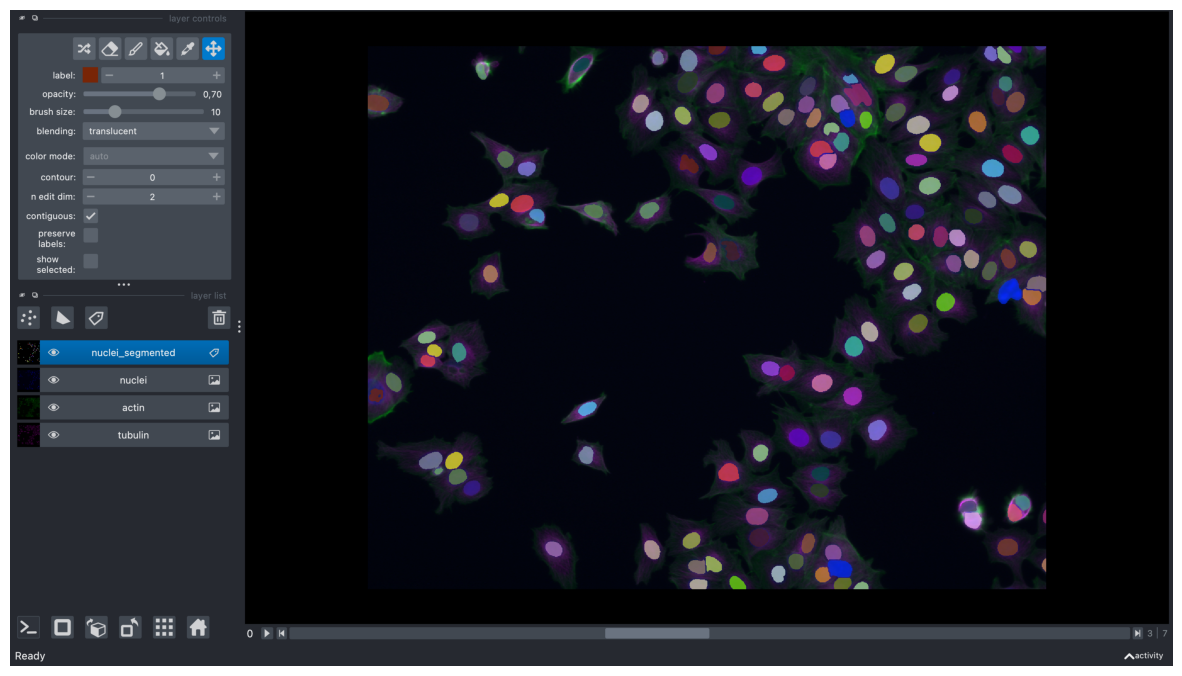
viewer.add_labels(labels_timelapse, name="nuclei_segmented")
<Labels layer 'nuclei_segmented' at 0x1777c60d0>
# note: some layers visibility and/or contrast limits were adjusted interactively in the viewer
screenshot = viewer.screenshot(canvas_only=False)
plt.figure(figsize=(15, 10))
plt.imshow(screenshot)
plt.axis('off')
plt.show()
save_path = os.path.join(path, "labels_nuclei")
if not os.path.exists(save_path):
os.makedirs(save_path)
tif.imwrite(os.path.join(save_path, f'{filename}_labels_dapi.tif'), labels_timelapse)
Process a batch of images from a folder#
save_path = os.path.join(path, "labels_nuclei")
if not os.path.exists(save_path):
os.makedirs(save_path)
for filename in tqdm(os.listdir(path)):
if not filename.endswith("tif"):
continue
# print(f"Processing image {filename}")
rgb_image = tif.imread(os.path.join(path, filename))
# take the blue (DAPI) channel only
img = rgb_image[:, :, :, 2]
# initialize an empty list where we will put obtained label images
labels = []
# iterate though the "time" dimension of the image
for t in range(img.shape[0]):
# gaussian blur
blurred_image = cle.gaussian_blur(img[t], None, 2.0, 2.0, 0.0)
# remove background
bg_removed = cle.top_hat_sphere(blurred_image, None, 20.0, 20.0, 0.0)
# threshold otsu
binary_nuclei = cle.threshold_otsu(bg_removed)
# split touching objects
binary_nuclei_edited = nsbatwm.split_touching_objects(binary_nuclei, 4.0)
# additional erosion to split a few still touching nuclei
binary_eroded_nuclei = binary_erosion(binary_nuclei_edited, disk(2))
# connected component labeling
labelled_nuclei = nsbatwm.connected_component_labeling(binary_eroded_nuclei, False)
# expand eroded nuclei back
dilated_labels = morphology.dilation(labelled_nuclei, disk(2))
labels.append(dilated_labels)
# we want to have only unique objects in each timepoint because it is not an actual timelapse!
# adjusted_labels_array = adjust_labels(labels)
# stack all obtained label images to a "timelapse"
labels_timelapse = np.stack(labels, axis=0)
# save the labels image
tif.imwrite(os.path.join(save_path, filename.split(".tif")[0] + "_labels_dapi.tif"), labels_timelapse)
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 17/17 [00:40<00:00, 2.41s/it]
